Click on a topic below for more detailed information:
Projects
We are well-seasoned gene jockeys, just ask and chances are we can clone it.
We routinely take on small (<10 samples) and large (48 or 96 samples) scale projects. Please see our price sheet to get an idea of our rates. Don’t hesitate to contact us with any questions.
We routinely use LIC (ligation independent cloning) and SLIC (sequence and ligation independent cloning) to make our plasmids. We can clone your gene into your plasmid or one of the many bacterial or baculovirus expression plasmids we have available. We also can synthesize custom vectors to provide the exact sequence you desire.
Click here to download our expression vector lists (includes a summary of all our plasmids, full plasmid sequences, and our cloning protocols)
All of the MacroLab Vectors are available at Addgene.org. For UC Berkeley labs, contact us directly.
If you are interested in obtaining our entire collection (as of DEC 2013) of 135 cloning plasmids please contact Scott Gradia. The two-plate set of glycerol stocks is available to UC labs for $900 and non-UC labs for $1400.
If we don’t have a vector that suits your need, it is likely that we can use SLIC or traditional restriction enzymes to clone into any vector you want.
More information about LIC cloning protocol here.
Helpful downloadable files:
- Easily design primers with LIC compatible ends using the following spreadsheet. Just enter the open reading frame that you want to amplify. Automated PCR Oligo Design
- Use the following spreadsheet to obtain the amino acid sequence, molecular weight, extinction coefficient, and PCR primers from your DNA sequence. MacroLab Molecular Bio Calculator
Please ask for guidance before you sit down and design your 96 well project. Hopefully we can save you a lot of time and effort.
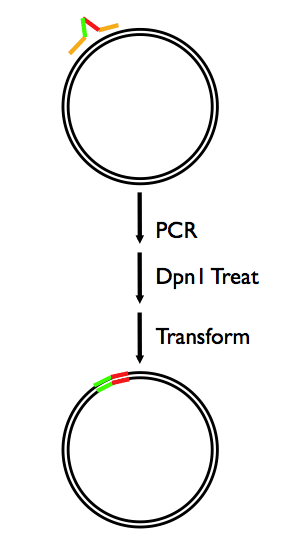
We have a very robust, one-oligo, method for quick-change mutagenesis.
We routinely perform 24, 48, or 96 reactions for our customers. We pick 4 colonies from each sample, then miniprep for sequencing in 96-well plates.
If your looking to make larger insertions or deletions, we have found that a SLIC method gives a higher success rate.
Quick Change Mutagenesis Protocol
Primer Design
Design one forward primer to anneal at 55C both upstream and downstream of the point mutation (approximately 45bp total primer length). Use the primer design spreadsheet if you need it. Change the fewest number of bases possible to get the desired amino acid.
Mutagenesis Reaction
| Volume (ul) | Reagent |
|---|---|
| Volume (ul)4 | ReagentPlasmid (160ng total) |
| Volume (ul)4 | Reagent1uM primer |
| Volume (ul)0.16 | ReagentdNTPs (100mM) |
| Volume (ul)0.4 | ReagentPfu Ultra* |
| Volume (ul)2 | ReagentPfu Ultra buffer |
| Volume (ul)9.44 | ReagentH2O |
| Volume (ul)20 total | Reagent |
* NOTE: It is very important to use Pfu Ultra, NOT Pfu UltraII!!
Cycle At:
| 95C | 2min | ||
|---|---|---|---|
| 95C | 95C | 2min20sec | |
| 95C | 53C | 2min1min | 18 cycles |
| 95C | 68C | 2min2min/kb up to 20min | |
| 95C68C | 2min1min |
DpnI Digest
| Volume (ul) | Reagent |
|---|---|
| Volume (ul)4 | ReagentMutagenesis reaction |
| Volume (ul)2 | ReagentNEB Buffer 4 |
| Volume (ul)0.5 | ReagentDpnI |
| Volume (ul)13.5 | ReagentH2O |
| Volume (ul)20 total | Reagent |
Incubate at 37c for 2.5-3 hours.
Anneal & Transform
1. Add 6ul to 100ul of competent cells on ice
2. Incubate on ice for 30min
3. Heat shock at 42C for 30sec
4. Immediately place on ice for 2min
5. Add 900ul 2YT and shake for 1hr at 37C
6. Plate 150ul on appropriate antibiotic plate
For a 96-well project, use the spreadsheet below to easily design your primers.
96 QC Primer Worksheet
You can also download the quick change mutagenesis protocol here.
Bacterial expression of your clones is performed in 96-well format. We use RIPL or Rosetta2 cells in auto-induction media (In Vitrogen Magic Media) for expression testing. We then use the Promega Magnehis Ni beads to perform automated Ni pull-downs on the Biomek3000. Protein gels of the soluble extract and Nickel Purification are run on our Caliper GXII.
We routinely perform E.coli expression and purification services for our customers using our AKTA Explorers and AKTA Xpress 4-pack. Our standard purification protocol uses nickel purification of a target protein that contains a Tev-protease cleavable His6 tag. A “subtractive” or “ortho” nickel purification is performed after TEV cleavage of the His6 tag as shown in the diagram below.
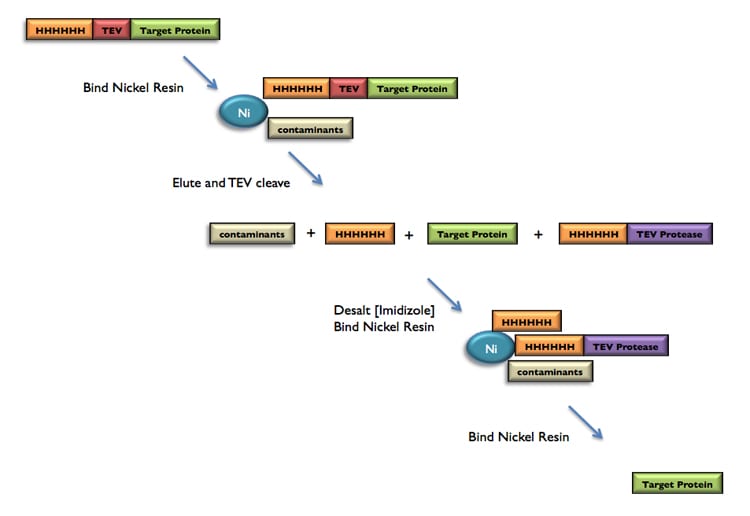
Depending on the purity of the target protein at this point, we may perform an ion exchange purification step before performing a gel filtration step. The target protein’s concentration, buffer conditions, and aliquot size are adjusted to meet your needs.
Please contact us for more information.
The MacroLab provides TEV protease for our in-house users. We put considerable effort into comparing various TEV expression clones and purification methods so that we can provide you with affordable, yet high quality TEV. We test each batch with our own TEV assay and we are confident it will provide you with robust and reproducible results.
TEV Information
Brief Methods
We express a double mutant (L56V / S135G ) pRK793 plasmid in Rosetta2(DE3)pLysS cells. 2L of cells are purified on a HisTrap Ni column, and the product is desalted with a 26/10 column. Final purification is performed on a HiTrap Capto-S column. Typical purity is shown below:
Your TEV Protease
*Keep your TEV frozen at -80C*
Your box of TEV contains 40mg of protein. We standardize the TEV concentration to 2mg/mL and aliquot each batch into 80 tubes. Each tube contains 250uL (0.5mg). Remember, unless you requested otherwise, the TEV is His6 tagged (so you can perform a subtractive nickel column with it).
Tev is frozen in the following:
25mM HEPES pH 7.5
400mM NaCl
10% glycerol
1mM DTT
We recommend cleaving your protein for at least 2 hr at room temperature or overnight at 4oC with a 1:20 (weight/weight) TEV/substrate ratio, or a 1:50 molar ratio. Some substrates may require more TEV. TEV is not inhibited by PMSF (1mM) , pepstatin A (1mM) , or complete protease inhibitor cocktail (Roche).
Each batch of TEV we produce is tested and known to cleave >90% of a substrate protein at a molar ratio of 1:50 in 2 hr at room temperature.
Special Handling Instructions
*Keep your TEV frozen at -80C*
TEV is prone to aggregation, so use it as soon as possible after thawing.
This is the protein expressed by TEV-DM-Prk793
MKIEEGKLVIWINGDKGYNGLAEVGKKFEKDTGIKVTVEHPDKLEEKFPQVAATGDGPDI IFWAHDRFGGYAQSGLLAEITPDKAFQDKLYPFTWDAVRYNGKLIAYPIAVEALSLIYNK DLLPNPPKTWEEIPALDKELKAKGKSALMFNLQEPYFTWPLIAADGGYAFKYENGKYDIK DVGVDNAGAKAGLTFLVDLIKNKHMNADTDYSIAEAAFNKGETAMTINGPWAWSNIDTSK VNYGVTVLPTFKGQPSKPFVGVLSAGINAASPNKELAKEFLENYLLTDEGLEAVNKDKPL GAVALKSYEEELAKDPRIAATMENAQKGEIMPNIPQMSAFWYAVRTAVINAASGRQTVDE ALKDAQTNSSSNNNNNNNNNNLGIEGRGENLYFQ^GHHHHHHHGESLFKGPRDYNPISST ICHLTNESDGHTTSLYGIGFGPFIITNKHLFRRNNGTLVVQSLHGVFKVKNTTTLQQHLI DGRDMIIIRMPKDFPPFPQKLKFREPQREERICLVTTNFQTKSMSSMVSDTSCTFPSGDG IFWKHWIQTKDGQCGSPLVSTRDGFIVGIHSASNFTNTNNYFTSVPKNFMELLTNQEAQQ WVSGWRLNADSVLWGGHKVFMVKPEEPFQPVKEATQLMNRRRRR^GHHHHHHHGESLFKG PRDYNPISSTICHLTNESDGHTTSLYGIGFGPFIITNKHLFRRNNGTLVVQSLHGVFKVK NTTTLQQHLIDGRDMIIIRMPKDFPPFPQKLKFREPQREERICLVTTNFQTKSMSSMVSD TSCTFPSGDGIFWKHWIQTKDGQCGSPLVSTRDGFIVGIHSASNFTNTNNYFTSVPKNFM ELLTNQEAQQWVSGWRLNADSVLWGGHKVFMVKPEEPFQPVKEATQLMNRRRRR
While it is expressing, self cleavage removes the N-terminal MBP and you are left with:
GHHHHHHHGESLFKGPRDYNPISSTICHLTNESDGHTTSLYGIGFGPFIITNKHLFRRNN GTLVVQSLHGVFKVKNTTTLQQHLIDGRDMIIIRMPKDFPPFPQKLKFREPQREERICLV TTNFQTKSMSSMVSDTSCTFPSGDGIFWKHWIQTKDGQCGSPLVSTRDGFIVGIHSASNF TNTNNYFTSVPKNFMELLTNQEAQQWVSGWRLNADSVLWGGHKVFMVKPEEPFQPVKEATQLMNRRRRR
pI 9.59
MW 28573.44
L56V and S135G improve protein solubility and thermal stability S. van den Berg, J. Biotechnol., 121 (2006), pp. 291-298
S219V – makes protein stable against autolysis. R.B. Kapust, Protein Eng., 14 (2001), pp. 993-1000
Download the TEV Info Sheet
Rosetta2 (DE3) pLysS
We stock:
- S.pyogenes Cas9: wild-type and high-fidelity (R691A) variant.
- AsCas12a Ultra.
We can purify a variety of other Cas9 variants on request. Contact us for details.