A new study in science shows how electron cryo-microscopy (cryo-EM) can shed light on the molecular details behind the gene silencing mechanisms that power cell differentiation.
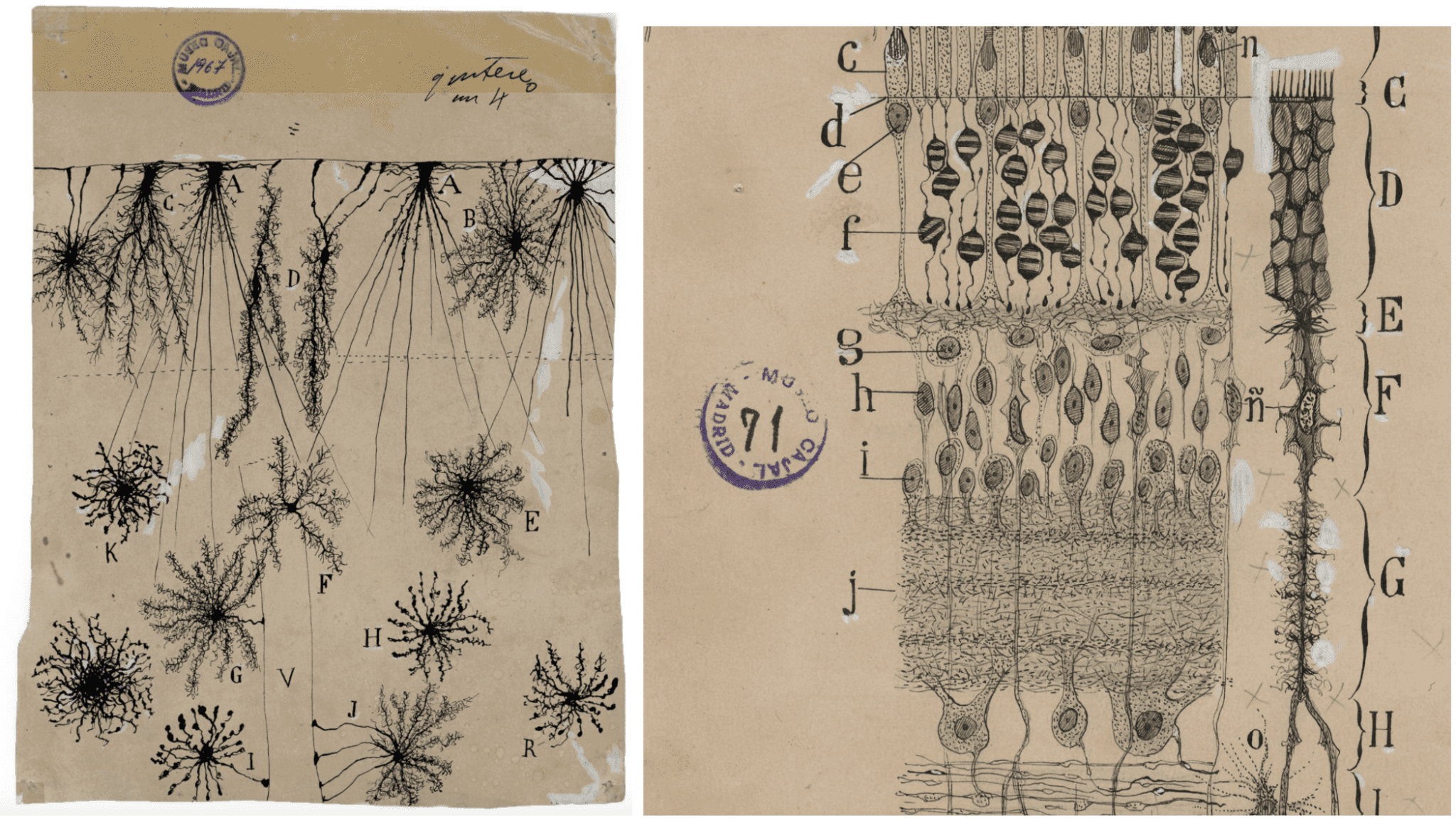
As a child growing up in the mid-19th century, Santiago Ramón y Cajal dreamt about becoming an artist. His father, a medical doctor, wouldn’t encourage his son’s artistic ambitions, so Cajal changed course and turned to science, ultimately becoming one of the most influential Spanish scientists in history.
Cajal’s (1852-1934) expertise was in neuroanatomy and the histology of the nervous system. He spent hours sketching detailed drawings of the different cell types he observed when looking at the human brain under a microscope, as well as the connections that brought these cells together to form tissues.
Looking at his drawings, a natural question to ask is how the enormous difference in cell morphology, that hints to functionally distinct roles, arises when virtually all of the DNA in each individual human cell is identical. The key lies in what we now call epigenetics, the study of diverse chemical marks on the DNA and associated proteins that can turn specific gene programs on and off and contribute to cell differentiation, the process by which cells become specialized. A laboratory led by another Spanish scientist, Eva Nogales at the University of California, Berkeley, has been studying these processes at the molecular level for years.
PRC2: a master key for turning genes off
In the article published today in Science, researchers from the Nogales lab employ a combination of electron cryo-microscopy (cryo-EM) and biochemistry to reveal the molecular gymnastics between proteins that turn genes off, ultimately contributing to determining the fate of cells. Building on previous studies, the team has been able to visualize these processes with an unprecedented level of detail.
For a stem cell to differentiate and become, say, a neuron, certain genes need to be inactivated. This is where molecular complexes in the cell nucleus jump into the picture. Major players regulating this process can be found in a family of proteins known as polycomb repressive complexes (PRCs). One of these biomolecules is a protein complex called PRC2, which is involved in the remodelling of DNA and the silencing of genes all across biology, from plants to humans.
In the fruit fly, for example, PRC2 helps to define the early “map” of development—the places where the different organs and structures, like legs and wings, will start to form—by silencing—or turning off—a gene named HOX in certain parts of the embryo but not in others.
The PRC2 complex also affects the way in which our own cells “make decisions”. “The DNA in our cells is wrapped around specialized proteins called histones,” says Vignesh Kasinath, the first author of the paper published in Science. “Together they form packages that you can imagine as the structural units of DNA in humans and all other eukaryotes. These small packets are the nucleosomes, and chromatin is formed by having many of these packets combined together.”
Throughout evolution, cells have tuned unique mechanisms to “paint” these histones in the nucleosomes with a variety of chemical signals. This set of chemical modifications can be combined in different ways across the genome, giving rise to complex patterns in each cell and leading to diverse outcomes. This is similar to how a person driving a car can integrate multiple queues from their environment—traffic lights, stop signs, pedestrians walking, etc—to make decisions. PRC2 decorates one histone in the nucleosomes with one particular “color”— a tri-methylation, which is a small chemical group.
There are several reasons to find out more about the regulatory mechanisms by which this family of gene-repressing complexes recognizes and modifies chromatin. For a start, polycomb repressive complexes are crucial in the physiological process of embryogenesis, where cells activate and repress various gene programs that drive their differentiation into specific cell types.
PRC2 also plays a major role in maintaining this cell identity once it has been established. In proliferative diseases like cancer, cells lose the tight regulation that arises from the symphony of molecular complexes orchestrating which genes are turned on and off. This eventually leads to “selfish” cells that divide uncontrollably and without an emergency brake.
“PRC2 is completely misregulated in cancer,” says Nogales. “The cells no longer remember whether they are lung, or uterus, or prostate cells. They reproduce in an uncontrollable way without any functional role.” PRC2 is associated with many different types of cancer, where mutations anywhere across the interface between nucleosome and PRC2 might lead to a misregulation in shutting down some genes and allowing cells to lose control.
By regulating which nucleosomes are marked with the tri-methylation—this chemical repressive signature—the cell can fine-tune which genes are turned off at different points in time. One of the major questions in the field of repressive complexes lies in understanding how PRC2 “decides” where to introduce these repressive chemical marks to take specific genes out of business.
For some time, scientists have known that another member of the polycomb repressive family, PRC1, introduces a ubiquitin mark—another chemical modification— into a different histone in the nucleosome. Evidence from previous work suggested that this chemical group could act as a lighthouse to recruit PRC2 and turn off the genetic information written in that nucleosome. One of the major breakthroughs of this work was to image the PRC2 complex in the context of a ubiquitinated nucleosome using a technique called electron cryo-microscopy (cryo-EM).
Shooting electrons through ice
“My lab has been using cryo-EM for many years, but now it has become very popular and extremely powerful at visualizing molecules,” says Nogales, the senior scientist leading the study.
But why is visualizing molecules so important to understand what makes them tick? “Humans are very visual animals, so having a visual representation can tell us a lot”, says Nogales.
Biological objects in the nanoworld have roughly the size of a millionth part of a millimeter, so visible light can not be used to observe these molecules in detail, like in an optical microscope. Fortunately, electrons have physical properties that make them suitable for accessing atomic information of complexes like PRC2 by imaging them under extremely cold conditions.
By rapidly freezing a purified sample containing many copies of the molecules to visualize, a thin layer of vitreous ice traps these complexes in close to native conformations and, ideally, in random orientations. This frozen sample is then transferred to an electron microscope that shoots high-energy electrons that are scattered by the atoms in the biomolecules and form an image at the detector. But how can we reconstruct a 3D structure of an object from looking at 2D images? According to Nogales, “each 2D image actually contains information on the whole 3D object and this is because the images are what we call projections. You can think of this as the density in the protein being collapsed, or squeezed into 2 dimensions. The structural information is there but we need a clever way to recover it.” Thanks to powerful algorithms that have been improved in the last decade, the information can be put together to produce a 3D map of the density in the protein.

Silencing genes with a little help from cofactors
Like most proteins, PRC2 does not act alone. This protein complex has a modest activity on its own and needs other protein cofactors that can aid in recognizing and modifying the regions of the genome that need to be repressed. “New cofactors for PRC2 are being discovered quite often,” says Kasinath.
We can think of these auxiliary proteins as tools in a Swiss army knife that impact the activity of PRC2 in different ways. The recent study from the Nogales lab focused on two particular cofactor proteins that have shown essential for early stages of stem cell differentiation—where a cell that doesn’t have a specialized role acquires one—JARID2 and AEBP2. Previous studies were showing that these two proteins play a role in recruiting PRC2 to nucleosomes, but little was known about the chemical and structural details of how this process was taking place. The Nogales lab found that ubiquitination—the epigenetic mark that “lights up” the nucleosome to be recognized by PRC2— of the nucleosome by PRC1 and recruitment of PRC2 are coupled, and that it is mediated by the two cofactors in a quite surprising way. This Swiss army knife proteins bind to opposing faces of the nucleosomes, breaking its characteristic symmetry, while also interacting with ubiquitin and PRC2. These multiple interaction interfaces enhance the binding of PRC2 to the nucleosome likely by reducing the rate at which PRC2 dissociates from the nucleosome. “We showed that in the presence of both cofactors and a nucleosome that has ubiquitin, PRC2 becomes a much better enzyme,” notes Kasinath.
Future structural work on this fascinating complex will help us better explain how the repressive signals are propagated across the genome and how other epigenetic marks can put a break on PRC2 and prevent necessary genes from being silenced.
Looking at the final frontier of what structural biology can offer today, the global cryo-EM community is pushing the limits of the technique to determine the structure of biological machines like PRC2 inside the cell. This would allow scientists to understand how PRC2 behaves in its native environment in health and disease, which might show some differences to what we observe in a purified sample.
These advances in structural biology are opening up an unexplored window into the molecular details of how the different cell types that Cajal elegantly portrayed in his sketches can naturally emerge from cells that have essentially the same information. At the pace at which cryo-EM technologies are moving forward, the realm of possibilities looks endless.
Co-authors of the paper include Vignesh Kasinath, Curtis Beck, Paul Sauer, Simon Poepsel, Jennifer Kosmatka, Marco Faini, Dan Toso, Ruedi Aebersold and Eva Nogales.