Graduate student John Berude highlights a new discovery from the Portnoy lab at UC Berkeley that helps scientists understand how food-borne pathogens know when they’ve reached their target.
UC Berkeley scientists have uncovered a new way that pathogenic bacteria might “know” when they have reached their desired destination: you.
The research, published recently in PLoS Pathogens, implicates host-derived methylglyoxal as a signal to the food-borne pathogen Listeria monocytogenes that it has successfully reached its new niche, and that it’s time to up-regulate its virulence genes and make itself at home.
You likely only know this microscopic bacterium from the small placard in the grocery store next to an empty shelf: “Item recalled due to contamination by Listeria monocytogenes.” But this microbe deserves far more recognition than that. L. monocytogenes is the causative agent of listeriosis in humans and animals, and infection occurs when we consume contaminated foods. Though most of us will only experience mild gastrointestinal issues upon infection, these bacteria cause severe disease and death in pregnant and immunocompromised individuals, such as the elderly, with mortality rates of up to 30%. Understanding these pathogenic microbes, and how they sense their surroundings, is crucial for protecting these vulnerable populations.
When we need to navigate the physical world around us, we rely on our five basic senses: taste, touch, smell, sight, and hearing. But microbes don’t have the luxury of these same senses. Even tiny organisms like bacteria still need to find food, protect themselves from danger, and adapt to an ever-changing environment. While studying bacterial sensing mechanisms is inherently important, the situation becomes more urgent when we are studying pathogenic bacteria that cause disease in humans. In the ever-evolving field of microbial pathogenesis, our understanding of the environmental cues that drive pathogenesis is just now coming into focus. And it turns out that these cues are coming from us.
Luckily, scientists have been working to understand these cues, and they have a big discovery to share. “This is just very exciting,” says Andrea Anaya-Sanchez, the lead author on the publication and a PhD student in Daniel Portnoy’s lab at UC Berkeley.
In this recent study, the authors identified a new gene in L. monocytogenes that is critical for these bacteria to successfully infect human and animal hosts. This gene encodes an important enzyme that is responsible for detoxifying a poisonous molecule that is made by infected host cells. “It’s a new finding in listeria for sure,” says Portnoy, who has been running the lab at UC Berkeley for over 30 years.
But both Anaya-Sanchez and Portnoy will tell you that the most exciting part of this work is not the discovery of the gene itself, but the implications it has for bacterial pathogenesis. “This seems to be part of how listeria activates its virulence program,” says Portnoy. “We think that the host is producing this molecule and listeria is using it as an indicator that they are in a host, and this is what actually activates their virulence program.”
The life of a pathogen
The biphasic lifecycle of L. monocytogenes is intriguing. Most of the time, the bacteria lead simple, harmless lives out in the environment, growing on decaying plant matter. However, everything changes when the bacteria meet human and animal hosts when we ingest contaminated food products. Upon entering a suitable host, the bacteria invade our cells and begin to express virulence genes that allow them to thrive inside our bodies.
During infection, these microbes hide inside our own cells and ramp up production of proteins that enable them to replicate intracellularly, evade our immune system, and spread to other host cells. At the center of this orchestra is listeria’s master virulence regulator, PrfA. This important protein is the conductor of the virulence symphony. When the bacteria are growing in the environment, PrfA is inactive, patiently waiting to cue up the band. But when the bacteria encounter a host, PrfA leaps into action and induces expression of virulence factors that allow the bacteria to happily proliferate in their new niche. Perhaps one of the most impactful recent discoveries in the field was made by a previous postdoc in the Portnoy lab, Michelle Reniere. She found that PrfA activation is dependent on the availability of one small molecule: glutathione.
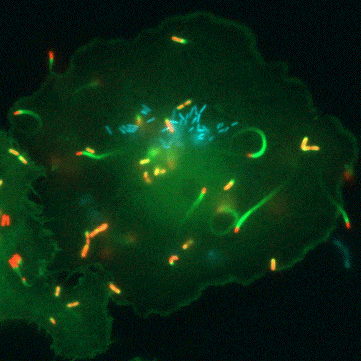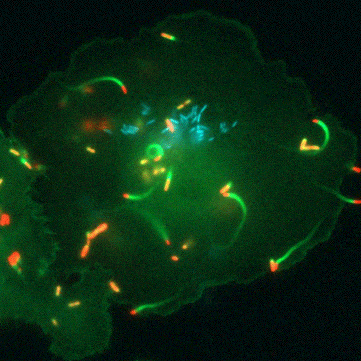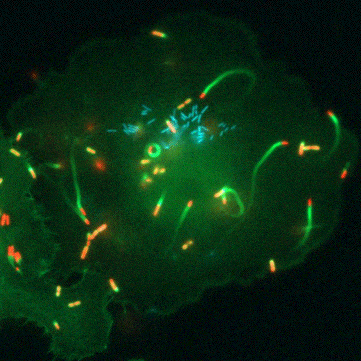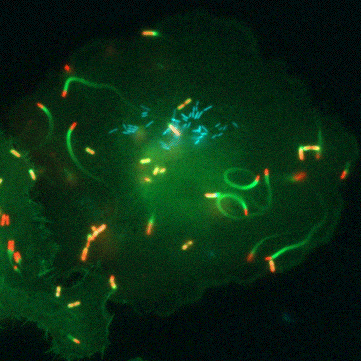
“Glutathione is a tripeptide that is present in all the cells of our bodies… and it’s a universal antioxidant,” says Portnoy. All life has a complicated relationship with oxygen. Even though we need oxygen to live, this fickle element of life is highly reactive and the cells in our bodies are constantly plagued by the havoc it can wreck. Oxygen can damage proteins, DNA, and even the very membranes that hold our cells together. This is where glutathione comes in. It’s a small, ubiquitous molecule that can clean up after oxygen. Because of this, all human and animal cells must produce large amounts of glutathione to stay one step ahead of this pervasive threat.
All cells that live in the presence of oxygen produce glutathione, or at least something like it, to combat oxygen’s damaging effects. “Some bacteria make glutathione, but the group of bacteria that Listeria belongs to doesn’t,” says Portnoy. So it was surprising when researchers discovered that L. monocytogenes has a special enzyme to make glutathione itself.
During infection, L. monocytogenes needs to activate its master virulence regulator, PrfA, and to do this, the bacteria need glutathione. Glutathione binds to PrfA, enabling the bacteria to express critical virulence factors and establish themselves in their host. But this glutathione must come from somewhere, and studies show that the bacteria can both import it from the host cell and make it themselves using their special enzyme, glutathione synthase. “Once inside of the host cell,” Anaya-Sanchez explains, “listeria can use the glutathione that is in the host. But we know that glutathione made by listeria is more important for PrfA activation.” These microbes are committed to doing it the hard way.
Microbial sense and sensibility
Ever since this important discovery, scientists have speculated that the key to understanding virulence gene regulation in L. monocytogenes lay in unearthing what induces glutathione production in the bacteria. According to Anaya-Sanchez, “one of the most upregulated genes upon cell entry is glutathione synthase.” This was a big clue, a clue that ultimately led to our poisonous molecule, methylglyoxal, and its role in pathogenesis.
Methylglyoxal is a reactive molecule, much like oxygen, that is formed naturally as a toxic byproduct of glycolysis. Glycolysis is a vital process that happens in all cells, which transforms the sugars we eat into energy for our bodies. If methylglyoxal is not readily detoxified, it can damage our proteins and DNA, much like oxygen does. When these damaged molecules accumulate in humans, they can cause numerous health conditions such as diabetes, aging, and neurodegenerative disorders.
Cue glutathione once again. Just as with oxygen, glutathione is necessary for preventing methylglyoxal from damaging proteins and DNA. When methylglyoxal is present, glutathione readily reacts with it, allowing for its permanent detoxification by the enzymes of the glyoxalase system. The most important of these enzymes is called GloA, which is the gene that Anaya-Sanchez discovered is important for virulence in L. monocytogenes. Through GloA and the glyoxalase system, glutathione is replenished and is available to react with more methylglyoxal. Without this system, glutathione is rapidly depleted, and cells succumb to methylglyoxal toxicity.
The initial finding of GloA in L. monocytogenes was itself exciting, but it had the authors scratching their heads: if methylglyoxal is regularly made by all cells, by both pathogenic bacteria and the hosts they infect, then why is it so important for pathogenesis?
While all cells undergo glycolysis and regularly produce small amounts of methylglyoxal, cells infected with pathogenic bacteria have been shown to produce very large amounts of this toxic molecule. According to Anaya-Sanchez, “methylglyoxal is also toxic to the bacteria, and so we think that this [increased production of methylglyoxal] could be a defense mechanism of the host.” During infection, host cells produce unusually high levels of methylglyoxal, which can enter and kill invading bacteria. A risky move for the host, given the risk of damaging itself, but one that typically plays out quite well. Most of the time.
But L. monocytogenes has evolved to be one step ahead of us. At the fundamental level, Anaya-Sanchez and her colleagues found that methylglyoxal exposure induces high expression of the glutathione synthase gene in L. monocytogenes. This, she explains, prompts the bacteria to produce higher levels of glutathione, the critical activator of PrfA and virulence. The immediate goal is to eliminate the toxic host-derived methylglyoxal, but the real win is producing all that extra glutathione to fuel the expression of virulence genes. “Together, our data suggest that methylglyoxal acts as a host cue that leads to glutathione production and activation of PrfA,” says Anaya-Sanchez. And just like that, these tiny bacteria have turned our defenses against us.

With Anaya-Sanchez’s new discovery, we now have a more complete picture of how pathogens might sense their animal hosts. For L. monocytogenes, methylglyoxal appears to be central to this picture. Host cells and bacteria alike always produce low levels of methylglyoxal, and this toxic compound is readily cleaned up by a small pool of glutathione. But microbes invade host cells, these cells produce more methylglyoxal to kill the bacterial intruders. In response, the bacteria produce more glutathione to defend themselves. Over eons of evolution, L. monocytogenes has learned that more glutathione means that they are in a host cell, and so they have made a direct connection between this increased glutathione and pathogenesis. It’s hard to deny the evolutionary elegance.
A needle in a haystack
But the road to discovery wasn’t always easy, says Anaya-Sanchez: “the initial [genetic] screen was very elaborate and long.” To identify bacterial genes that are important for virulence activation and pathogenesis, Anaya-Sanchez conducted an arduous genetic screen for bacterial mutants that failed to grow inside host cells but that could be rescued in a special strain of bacteria that contains a mutation in PrfA that bypasses the need for glutathione. This special screen is what scientists call a plaque screen, where cells are infected with L. monocytogenes in vitro and screened for smaller plaques that indicate a bacterial strain with a potential virulence defect.
The screen itself involved sifting through over 100,000 potential mutants and took over a year, and the characterization of GloA in L. monocytogenes and its exact role in pathogenesis took even longer. So long, in fact, that Anaya-Sanchez started the research as a visiting master’s student, completed this degree, and then worked as a technician for a year, and then finally published the work as a second-year PhD student in the department of Plant and Microbial Biology at UC Berkeley.
But it was all worth the wait to Anaya-Sanchez and Portnoy. As Anaya-Sanchez puts it: “the host cues that listeria senses inside the host cell to activate its virulence program has been a longstanding question in the field, and when Michelle Reniere discovered that upregulation of glutathione synthase and glutathione availability were required for full PrfA activation I think we got a little bit closer to understanding how listeria recognizes its intracellular location,” she notes. “So, what I think is really significant and exciting about this study is that we took a step closer to finding the answer.”
The path forward
Just as with any great discovery, scientists are left with more unanswered questions than we started out with. “We’re now interested in understanding how important these [methylglyoxal] detoxification systems are in other bacteria, like Mycobacterium tuberculosis,” says Anaya-Sanchez. M. tuberculosis is the causative agent of tuberculosis, and one of the oldest and most significant human pathogens of all time.
It is also painfully difficult to study, requiring long incubation times with special training and facilities to protect researchers from accidentally infecting themselves with the microbe. “We’re now in collaboration with the Sarah Stanley lab,” notes Portnoy, “trying to make a similar mutant in Mycobacterium tuberculosis to see if it also affects virulence similarly.” And as a co-mentored PhD student in both the Portnoy and Stanley labs, Anaya-Sanchez is well-situated to find out.
Still more questions remain on the listeria side of the table, though, and both Portnoy and Anaya-Sanchez are eager to follow up on them. “We are excited to study DNA damage derived from methylglyoxal exposure,” says Anaya-Sanchez, “and whether it’s killing the bacteria and what other defense or repair mechanisms listeria is using to withstand DNA damage.”
Portnoy adds: “One of the consequences of gloA mutation is that [bacteria] suffer more mutations [in vitro] because methylglyoxal damages DNA, so I’m very excited to see what happens in vivo.” The studies are already underway.
But Portnoy has other plans too: “The lab got started thirty years ago with a plaque screen, and since then there have been so many new methods and [gene] annotations.” Because of these advancements, through Anaya-Sanchez’s new plaque screen, “we found all kinds of new genes to work on.” Genes, he insists, that may hold more clues about how pathogenic bacteria make us sick, and how we can fight them. So, for the field of microbial pathogenesis, the future seems as bright as ever.