UC Berkeley research strives to understand the cell’s complex recycling program and how we can use it to treat diseases.
Our bodies are composed of trillions of cells—each one equipped with a microscopic recycling center known as the ubiquitin-proteasome system, or UPS. When proteins inside the cell need to be removed, the UPS recognizes, marks, and recycles these proteins.
Research conducted at UC Berkeley has revealed many molecular details of the complex recycling program that is the UPS. As we begin to understand more about the elegant ways in which our cells recycle, researchers strive for a new goal: leveraging the cell’s own recycling plant to treat disease.
What is the UPS?
In many ways each cell in our body is a miniature city. Within the bounds of the cell, there lies a hustle and bustle as thousands of proteins scurry from location to location carrying out all the processes of life. Some are tasked with a singular purpose, such as modifying a single molecule or carrying oxygen. Some collaborate on breaking down our breakfast and turn it into energy. Others cluster en masse to form molecular factories, taking in energy and cellular materials to allow the cell to build, to grow, and eventually, to divide.
Like any major city, the cell produces waste and over time even the sturdiest of our protein machines wear down. With over 20,000 machines working in the cell, waste can accumulate quickly. The cell can’t leave its trash on the curb for pick up or sort its trash and send it to a recycling plant. So, how does the cell process its waste?
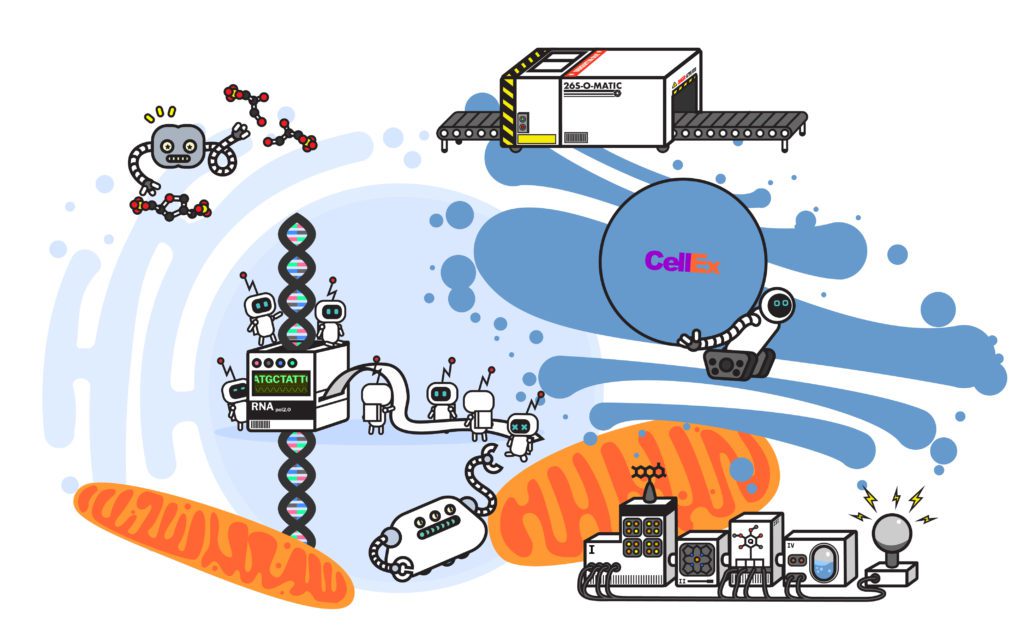
The cell handles the bulk of its waste using a cellular recycling program known as the ubiquitin-proteasome system. When a protein reaches the end of its life cycle, a network of enzymes, known as ubiquitin ligases, recognizes the faulty protein and tags it with another protein, ubiquitin. The ubiquitin protein is an extremely versatile signal, as it can take the form of a single-protein tag or a chain of multiple ubiquitins strung together. Often, the way the ubiquitin proteins are appended to each other gives them distinct shapes or branching patterns. Certain variations of these ubiquitin chains can act as a signal that the tagged protein needs to be recycled—ultimately picked up by one of the cell’s factories, the proteasome.
Mechanically, the proteasome is a protein processing plant. First it recognizes the ubiquitin tag as instructions and grabs the faulty protein. Then it carefully removes the ubiquitin tag—no need to let the ubiquitin go to waste—so it can be reused.
Finally, it uses a molecular motor to pry apart the faulty protein and disassemble it—the result: parts. The cell takes amino acids—the disassembled parts of old proteins—and reuses them to make new proteins.
This system of recognizing and recycling old, damaged, or simply not-needed proteins is both incredibly complex—it is estimated that ubiquitin ligases make up approximately 5% of the human genome!—and essential. Defects in the cell’s recycling system often result in severe consequences ranging from cancer to neurodegeneration.
Insights into the UPS from Berkeley scientists
Berkeley has long been a home for minds working towards improving our understanding of the UPS. Even before starting his lab here in 2006, Michael Rapé has been steeped in the ubiquitin field. As a postdoctoral scholar, Rapé worked on trying to understand the degradation events that occur as the cell prepares for the dramatic dance of mitosis, or cell division. Rapé discovered many facets of how a ubiquitin ligase known as the anaphase promoting complex (APC) helps the cell in making the decision to progress through mitosis.
“You have all these degradation events that happen at different times in mitosis— and it’s a very short time; mitosis, it’s less than an hour,” explains Rapé “and many of them are driven by APC, and the question was whether this was controlled by localization or by something else”.
He credits his dive into protein degradation to early work in his lab which showed that this degradative push into cell division by APC is driven by a specific ubiquitin signal, the K-11 ubiquitin chain, one previously not implicated in protein degradation. This work presented evidence for the idea that there is a kind of ubiquitin “code” inside the cell. Rapé explains that each different type of ubiquitin chain can act like a word and when you string them together you can make sentences. The cell can read these sentences and use the information to make decisions—such as if it’s an appropriate time to proceed with division. Rapé’s lab has continued to focus on translating and understanding these ubiquitin messages in a myriad of cellular decisions such as how large of a cargo to export out of the cell, what should the cell differentiate into, and when should the cell degrade one protein-protein interaction versus another.
Just up the hill in Stanley Hall, Andreas Martin contemplates the same system but from the side of the proteasome, the molecular factory that carries out the recycling. A veritable protein mechanic, Martin made a name for himself in the field of cellular protein motors by tinkering under the hood of the bacterial version of the proteasome, a molecular motor complex known as ClpX. It’s easy to see that Martin has a deep passion for understanding these intricate molecular machines—when asked about his transition from working on one motor to the next Martin states he saw the proteasome as “the next logical step, as the most complicated molecular motor complex.” He was excited to use its unique characteristics to better understand the motors that pull and push inside our cells.
When asked if he saw himself revolutionizing the way the UPS field views the proteasome, Martin smiles and responds with a resounding “no.” Yet, prior to Martin’s arrival in 2009, the architecture of this cellular factory was virtually undeciphered. Scientists often learn by breaking the systems they work on. Mutations and defects, causes and effects—they inform on form and function. The proteasome is so essential that mutations in its critical functions lead to death in the model organism, making it a difficult target for the learning-by-breaking strategy. Martin’s lab revolutionized the field by establishing a system to express proteasome recombinantly in bacteria, allowing them to make and test mutations that you could never make in a non-bacterial cell. In 2012, in collaboration with the expertise of Eva Nogales’ lab and leveraging the power of their recombinant proteasome system, Martin’s group presented the field with the first view of the layout of the complete proteasome, including the arrangement of receptors for ubiquitin recognition on the proteasome. “If you have a good structural framework,” notes Martin, “it really changes things—changes the way you think about systems”.
The Martin lab has continued to take cues from structural frameworks to paint a more complete picture of where the ubiquitin signal binds, how it is removed from a target, and the detailed mechanisms by which the proteasome unfolds proteins in the cell alone and in collaboration with other molecular motors. In 2018, the Martin group presented the first high-resolution structure of the proteasome caught in the act of unfolding a protein, once again providing atomic-level details into this essential cellular process. Over the years, basic scientific insights into the UPS, like those unearthed by the Rapé and Martin labs, have allowed researchers to begin wondering if the power of the UPS could be harnessed.
To degrade or not degrade: Using the cell’s waste disposal to treat disease
In 2001, a group of scientists, including Caltech professor and Berkeley PhD alum, Ray Deshaies, proposed a revolutionary idea: could it be possible to trick the cell’s waste management system into removing a protein it normally wouldn’t?
They reasoned that if one could artificially bring the waste-recognition machinery and a target protein together, perhaps this protein-of-interest would be tagged with ubiquitin and promptly shipped off for recycling by the proteasome. To do this, the team designed a chimeric molecule that would bind both a ubiquitin ligase and a protein-of-interest—one not typically targeted by the ligase. They named this molecule protein-targeting chimeric molecule 1 or Protac-1. Protac-1 was a success: it was able to recruit a ubiquitin ligase to degrade the protein MetAP-2, a completely new target for the ligase and an existing anti-cancer drug target.
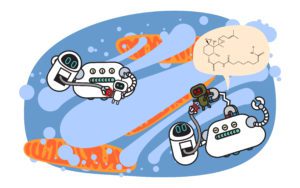
Since the publication of that original paper, “PROTAC” has transcended from the name of the original chimeric molecule into the callsign of an entire industry devoted to leveraging the cell’s waste recognition and recycling systems to treat disease. Several UC Berkeley investigators have taken up the challenge of targeted protein degradation, including Rapé, who started Nurix Therapeutics with colleague John Kuriyan in 2012, with the mission to expand the breadth of both the ubiquitin ligases and the proteins they target.
“In 2009 we formed Kura Therapeutics, which in 2012 was renamed into Nurix,” recounts Rapé. At the time, Kura was among the first companies focused solely on targeted protein degradation as a therapeutic. Nurix is now a publicly traded company with 250 employees across four different sites, but Rapé fondly recalls that Nurix at its inception was a fruitful collaboration between himself, Kuriyan, and Kyle Simonetta, a talented postdoc and former student of Kuriyan’s who began the earliest iterations of screens that would lay the groundwork for Nurix’s creation.
“What we do now—is not only to think about what the next great company would be,” explains Rapé, “but also where could you take the lessons learned from induced protein degradation to have really conceptually new compounds that treat disease.”
UC Berkeley’s Dan Nomura is not wasting time to do just that. In a recent preprint article posted to bioRxiv, the Nomura group describes a deubiquitinase targeting chimera, or DUBTAC, presenting an exciting proof-of-concept for a new paradigm in UPS-based therapy: targeted protein stabilization.
Akin to traditional PROTACs, the approach is to artificially recruit two proteins that normally would not interact. However, instead of recruiting a ubiquitin ligase to a target protein, DUBTAC recruits a ubiquitin-removing enzyme—a deubiquitinase or DUB—to a protein and prevents its degradation. Their results show that the strategy works to recruit a DUB, OTUB1, to stabilize a mutant of the CFTR protein. Abnormal forms of CFTR are recognized as faulty and promptly removed, but an unfortunate consequence of the UPS performing its job is that the removal of CFTR results in the onset of Cystic Fibrosis. Fortunately, prior work has shown that stabilizing CFTR—even in its mutant form—can alleviate the severity of the disorder. In the article, Nomura’s group screen for an OTUB1-binding molecule, chemically fuse it to an existing CFTR-binding drug and demonstrate their DUBTAC results in stabilization of the most common disorder-causing variant of the CFTR protein.
Nomura’s excitement about the future of DUBTACs is clear and his lab has sights set on other targets. “There are many other targets—hundreds of targets—where they’re actively degraded and there are ligands against those targets,” Nomura notes about the immediate future directions for DUBTACs. “So, those are lower-hanging fruit.”
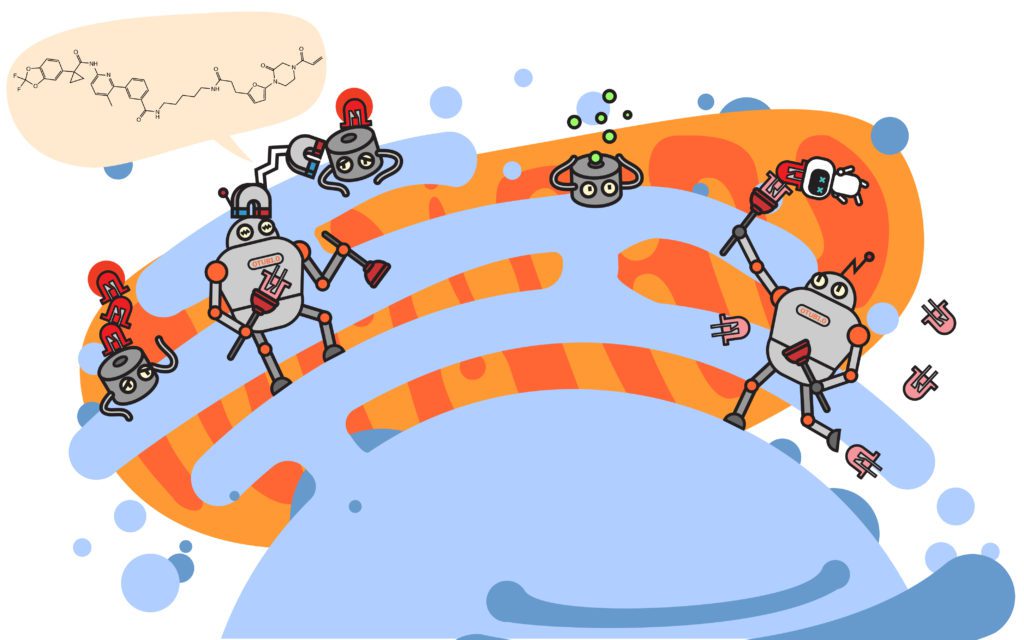
The ubiquitin-proteasome system didn’t evolve to treat cancer or degrade targets for Cystic Fibrosis, but insights from basic research has allowed researchers to co-opt an existing system to improve our health. New insights into the UPS and other cellular pathways are being garnered every year from research at UC Berkeley: A collaboration between the Susan Marqusee and Martin labs showed that the addition of ubiquitin and the position of ubiquitin itself can have major effects on the stability of proteins and their accessibility to recycling.
This year alone, the Rapé lab has uncovered roles for ubiquitination in the processes of cellular stress responses and coordinating cell fusion during muscle development and the Martin lab has recently taken a deep look at how the proteasome changes shape in response to ubiquitin binding, and how this may aid in degradation of cellular proteins. These discoveries may someday be used by scientists like Nomura who straddle the line between basic and translational research to transform our approaches to treatment.
“Berkeley tends to be basic-biology oriented,” says Nomura, “and our lab doesn’t shy away from the basic biology, but we’re very much focused on translating everything that we do into real-world applications such as therapeutics. Everyone in my lab has that goal—I have that goal. Every day I wake up, that’s the only thing I think about.”