The GSL now uses a secure encrypted FTP server (FTPS not SFTP) for data transfer. You will receive an email regarding login information. Please use an FTP client (e.g., FileZilla) or FTP from the command-line (e.g., lftp) that supports explicit FTPS. You may allow for multiple parallel connections to speed up downloading. Basic instructions for FileZilla and the command-line, as well as troubleshooting suggestions, are detailed below.
The GSL hosts data on our FTP server for 1 month following project completion. Raw data is kept on our live backup for a minimum of 1 month. Within this time fram, data can be requeed, reprocessed and hosted again upon request at no cost. Data more than 3 months old is moved to our long-term storage, AWS glaical storage. Retrieval from long-term storage will incur a processing fee ($1000).
Savio and Vector Users
Command-line lftp is the best option for transferring data to Berkeley’s computing cluster. You must be logged into a data transfer node (dtn.brc.berkeley.edu) and not a login node. See the Berkeley HPC user guide for further details.
1. Opening FileZilla, use the Site Manager to establish a secure connection. Do not use the Quickconnect functionality.
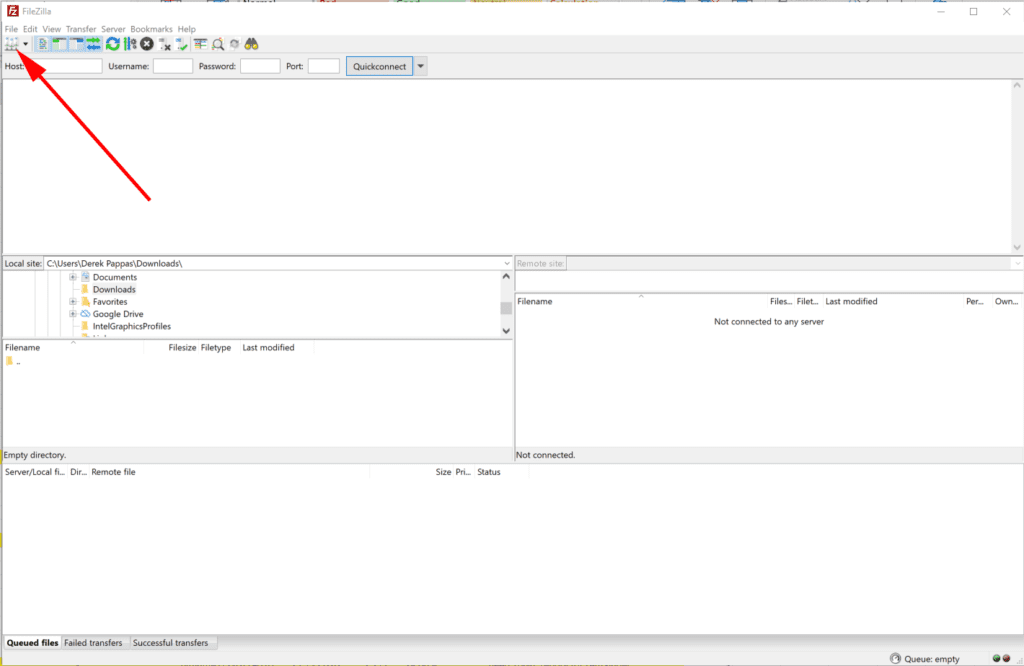
2. Fill in the relevant information as shown below substituting your login information in the user/password fields. Press Connect.
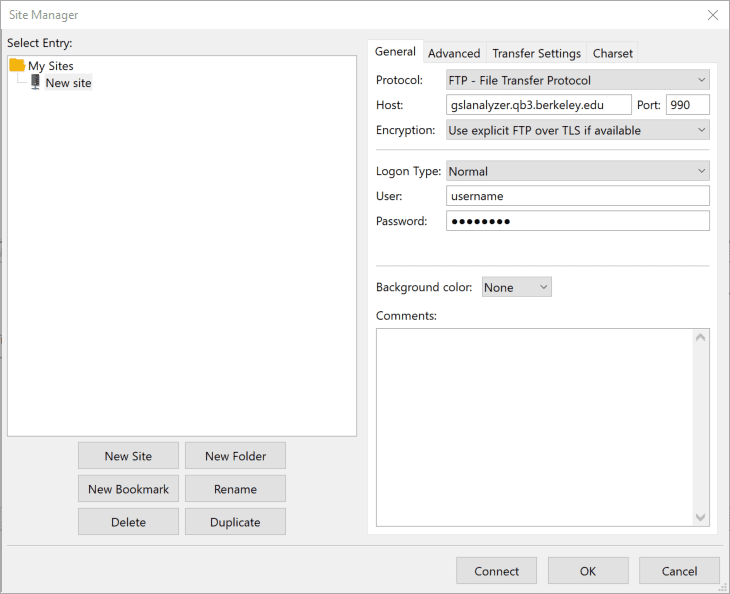
3. FileZilla will securely connect to the GSL server and display a directory listing of your files (Remote site pane, right). You can then select all your files (Ctrl+A) and drag them over to a local folder on your computer (Local site panel, left).
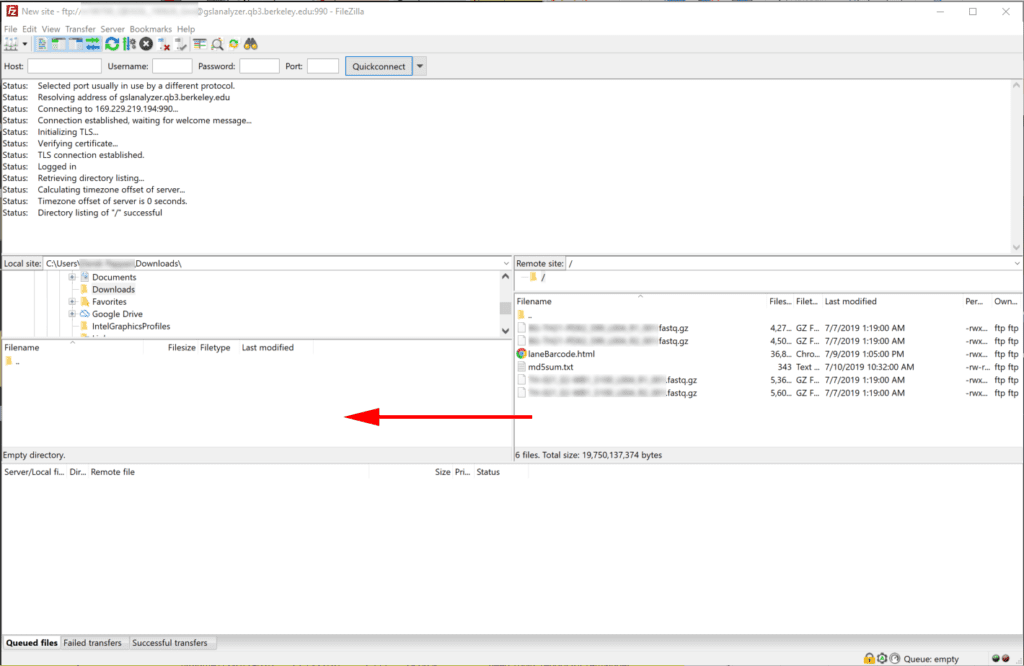
4. Downloading progress and success will display in the bottom and top panes respectively.
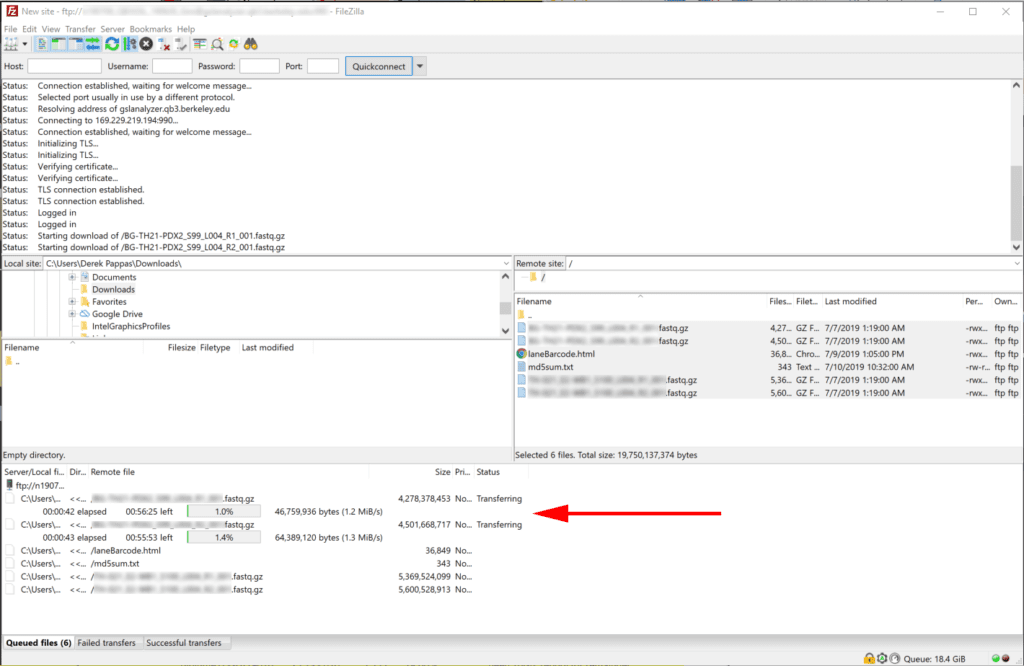
5. When all downloads have completed. The bottom panel will be empty and you may close FileZilla.
6. If you chose another ftps compatiable program (WinSCP, Cyberduck), please ensure similar settings.
1. lftp is a sophisticated command-line utility with many options. You can make a basic connection and log into your data directory using the following statement substituting your username/password from the “data available” email you received.
# … do not include command prompt $
$ lftp -u username,password -e “set ssl:verify-certificate no set ftp:ssl-protect-data true set ftp:ssl-force true” ftp://gslanalyzer.qb3.berkeley.edu:990
2. Assuming no error messages, you will be at the lftp command prompt. Use “help” (without quotes) to see a list of available commands. Common commands are “ls” to list files in your directory as well as “mirror” to retrieve files to your local directory.
# … lftp prompt after successful connection
lftp username@gslanalyzer.qb3.berkeley.edu
# … list files in directory
lftp username@gslanalyzer.qb3.berkeley.edu: ls
# … mirror files into local directory
lftp username@gslanalyzer.qb3.berkeley.edu: mirror
# … mirror files with continue option (recommended)
lftp username@gslanalyzer.qb3.berkeley.edu: mirror -c
# … mirror files with 10 parallel downloads to speed up transfer
lftp username@gslanalyzer.qb3.berkeley.edu: mirror -P 10
3. Once the downloads have completed, you may exit lftp using “exit/quit” at the prompt.
# … exit lftp
lftp username@gslanalyzer.qb3.berkeley.edu: exit
4. You can combine options in a single line command that skips the lftp prompt and downloads your files into your local directory (ensure standard straight single/double quotes).
# … do not include command prompt $
$ lftp -c ‘set ssl:verify-certificate no set ftp:ssl-protect-data true set ftp:ssl-force true; open -u username,password -e “mirror -c; quit” ftp://gslanalyzer.qb3.berkeley.edu:990’
5. If you chose another FTPS compatiable command-line option, please ensure settings for FTPS.
- Secure FTP connections: Our servers do not support SFTP. Please use an alternative such as FileZilla, lftp, wget, or curl that support FTP through explicit TLS (FTPS).
- Login incorrect: If you are receiving the login incorrect error, your password may have expired or requires regeneration. Please email the GSL and let us know.
- Secure FTP ports: If you are having connection troubles. The connecting/destination machine may need to open TCP ports 12000-12100, though generally this shouldn’t be necessary.
- Command-line errrors: Copy-and-paste from an email or this website may introduce formatting errors. For example, please ensure standard straight single and double quotes, not typographic curly opening/closing quotes when using command-line. Double hyphens can format as longer em dash on emails.
- FileZilla Quickconnect: Connecting to our secure servers via Quickconnect is not supported. Please use the FileZilla Connection Manager as detailed above.
- FileZilla savings passwords error: Ensure that saving of passwords through FileZilla main menu “Edit > Settings > Interface > Passwords” is checked and then retry the connection.
MD5 Checksums:
To assist in verifying the integrity of data downloads we provide md5 checksums for your fastq files. Each user may check their downloads against the md5sum.txt associated with their data. On Unix-based operating systems the command md5sum -c md5sum.txt will do this.