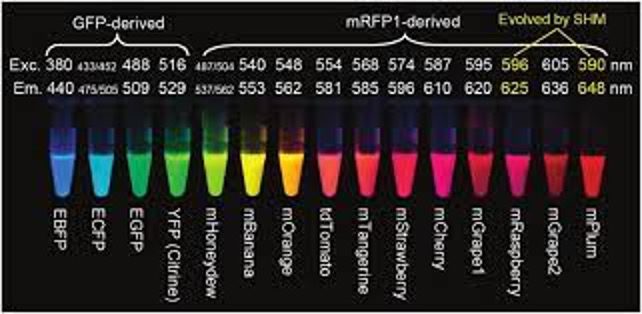
The discovery of GFP enabled many new applications of fluorescence imaging in biology. Researchers have developed a broad palette of mutant derivatives of GFP that absorb and emit at a range of different wavelengths. Along the way, they have optimized other characteristics of these fluorescent proteins, such as photostability, maturation time, and brightness. Other sources of fluorescent proteins were subsequently discovered in sea anemones such as Entacmaea quadricolor and the sea pansy Renilla reniformis, further expanding the range of options available for fluorescence imaging.
Ingenious derivatives of GFP have been developed whose fluorescence increases or decreases in response to changes in calcium concentration or membrane voltage. These genetically encoded fluorescent sensors make it possible to observe the firing of neurons in living brains using fluorescence microscopy.
We will observe the localization of a couple of proteins fused with GFP, which have been expressed in cultured mammalian cells. Mammals such as mice and rats are important model organisms in biology, as they are more closely related to humans than other organisms like yeast or flies. However, working with them is expensive and challenging for many reasons. Many biologists prefer to use cultured cells, as they are less expensive and more appropriate for their experiments.
In the 3T3 cells, GFP has been fused to heterochromatin protein 1 (HP1). In the nucleus, there are two broad classes of chromatin (packaged DNA): euchromatin and heterochromatin. In general, euchromatin contains actively transcribed genes, while genes in heterochromatin are repressed. HP1 is a major component of heterochromatin and is frequently used as a marker for this type of chromatin.
U2OS cells, originally derived from a human bone cancer called osteosarcoma, are commonly used for fluorescence imaging because of their large, flat shape. We will image U2OS cells that express GFP fused to a low complexity domain (LCD). LCDs are regions from proteins in which the amino acid sequence shows little diversity in composition, often due to long stretches of a repeated, short sequence. In particular, LCDs are present in many components of the transcription machinery, including sequence-specific transcription factors and RNA polymerase II. Studies have found that LCDs can interact specifically with each other. High concentrations of purified LCD-containing proteins can sometimes form gels or liquid droplets, and in cells, they may gather into large clusters that are visible under a microscope. In most cases, it remains unclear whether these types of interactions play a functional role, and this is an active area of research.
Slides with these cells have already been prepared for you to look at. In addition to expressing a GFP fusion protein, the cells have also been stained with a fluorescent dye called DAPI, which stains the DNA in the cells.
When it is your turn to use the microscope, find some cells and image both the GFP fusion protein and the DAPI stain with help from one of the instructors. After imaging, consider the following questions:
- How does the localization of the GFP fusion protein compare to the DAPI stain?
- How do the localizations of the GFP fusion proteins compare to each other?
- Is the localization of the GFP fusion protein similar in different cells in a given cell line? If not, what might be an explanation for this?